Announcement:

We are pleased to announce the introduction of GSA-BenchmarKING, a repository of GSA benchmarking tools:
GSA-BenchmarKING is a repository of apps/workflows/pipelines that allows benchmarking groups of GSA software in an easy and automated way. Currently, GSA-BenchmarKING contains jupyter notebooks with full workflows for benchmarking GSA methods, and shiny apps that allow benchmarking with the click of a few buttons.
Screenshots:
Fig 1. A jupyter notebook with a workflow for benchmarking single-sample GSA tools.
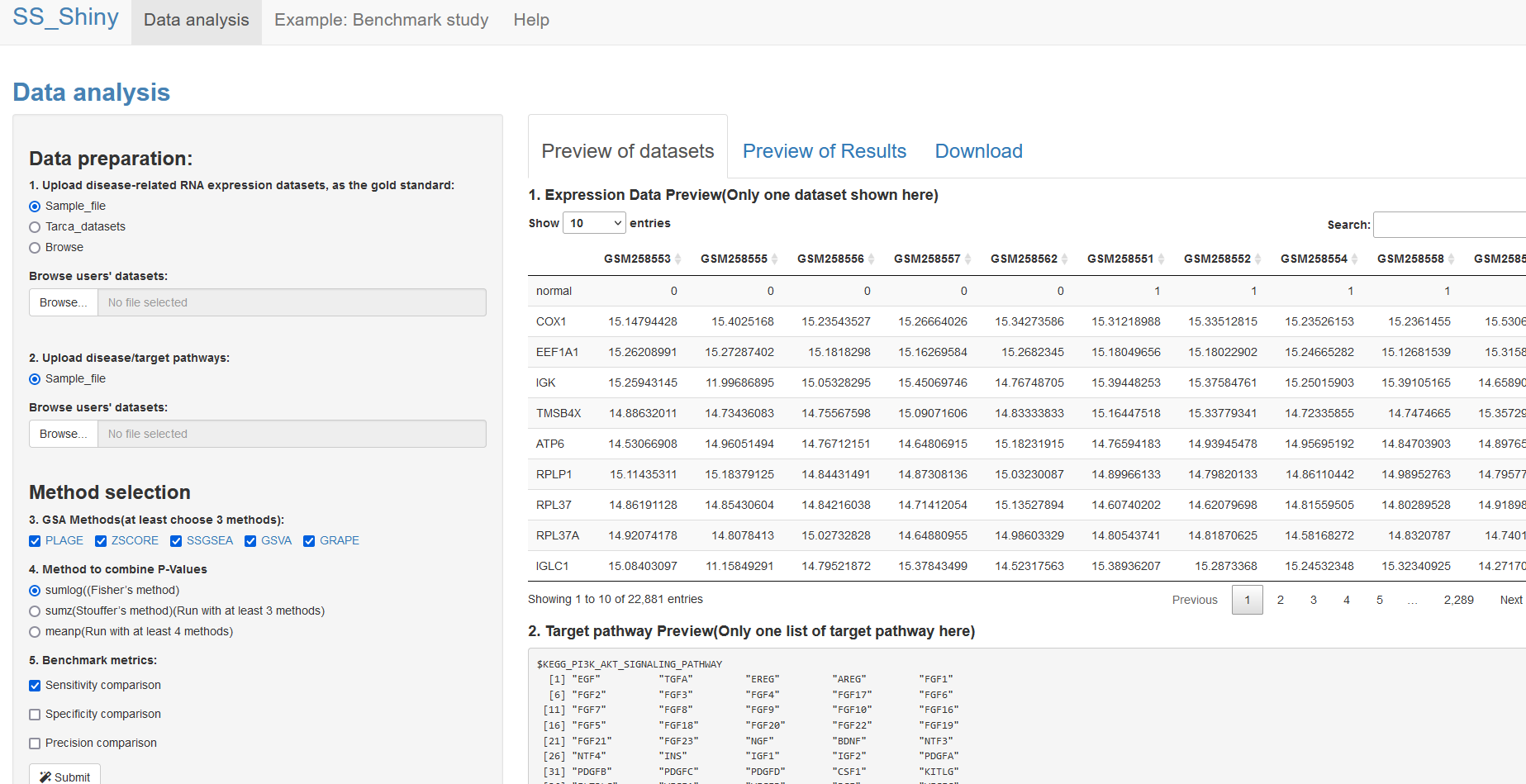
Fig 2. A shiny app for benchmarking single-sample GSA tools (dataset preview tab).
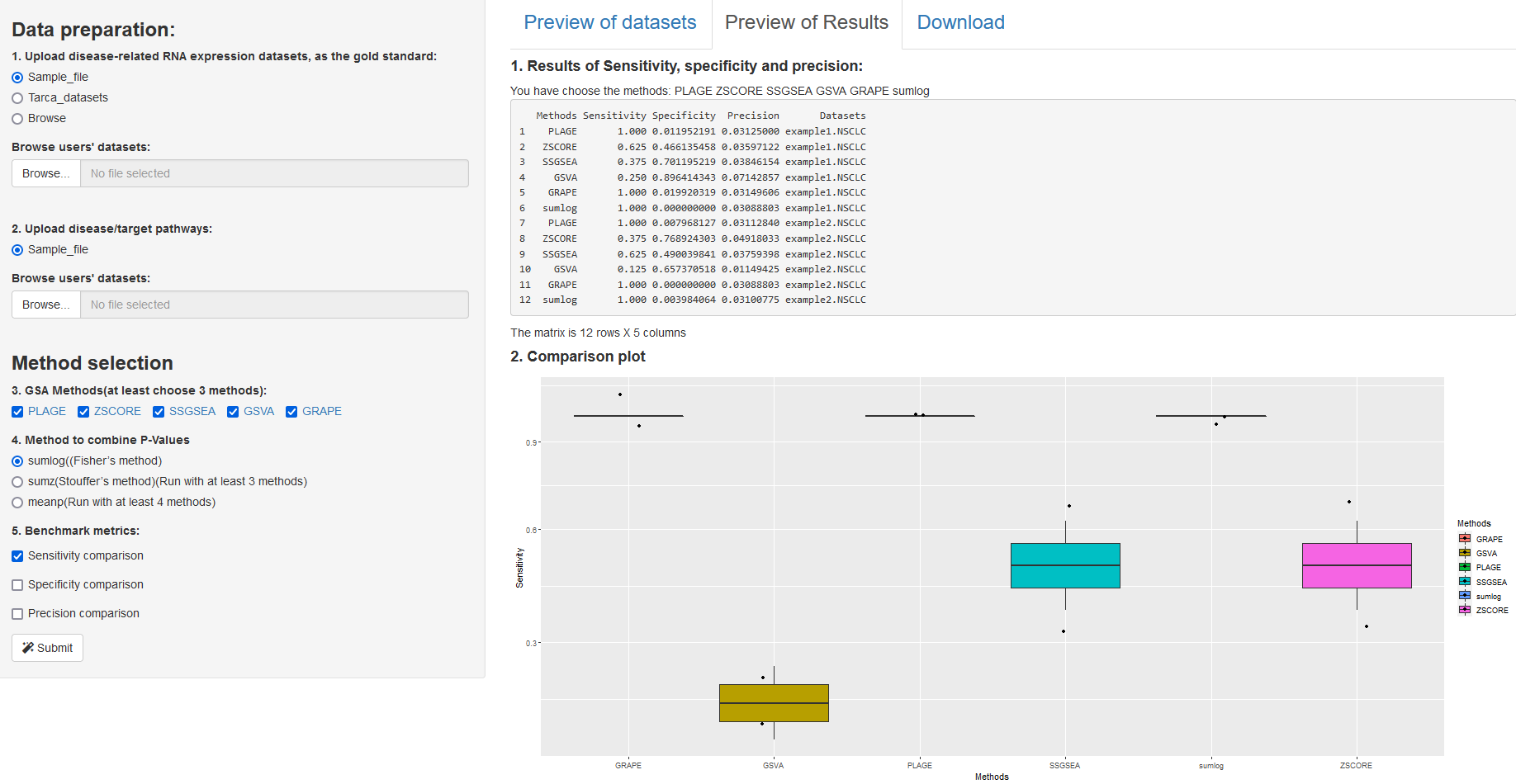
Fig 3. A shiny app for benchmarking single-sample GSA tools (results tab).
References:
GSA-BenchmarKING has been introduced in the following publication:
[1] XIE, C., et al. (2021), Popularity and performance of bioinformatics software: the case of gene set analysis, BMC Bioinformatics 22, 191, https://doi.org/10.1186/s12859-021-04124-5